Introduction: Nearly half of adults with acute lymphoblastic leukemia (ALL) will experience relapse of their disease, and those with relapsed ALL carry a worse prognosis than those who remain in remission. The influence of the tumor immune microenvironment (TME) on relapse and survival outcomes in ALL is not well understood. Mass cytometry (CyTOF) enables immune profiling based on single cell protein expression and can identify phenotypic diversity of cells in the TME that cannot reliably be detected by standard flow cytometry. Recently, T-cell/monocyte hybrid cells have been discovered in patients with uveal melanoma and latent tuberculosis infections. However, cells expressing CD19, a surface antigen expressed on B-ALL cells, and markers of other cells in the TME including T cells and tumor-associated macrophages (TAMs), have not been described in ALL or other hematologic malignancies. The presence of CD3/CD163 hybrid cells in the TME may have predictive and prognostic implications for adults with ALL, as TAMs play a role in suppressing antitumor immunity which can lead to T-cell exhaustion. In this study, we identify the presence of multilineage hybrid cells in peripheral blood (PB) and bone marrow (BM) samples of patients with B-ALL alongside accompanying evolution of the TME at each disease stage.
Methods: PB (N=16) and BM aspirate (N=16) isolates were collected from patients with B-ALL at diagnosis, relapse, or remission, as well as from healthy donors (HDs, N=7) for comparison of cellular profiles. A 28-marker CYTOF panel was designed to evaluate cell surface and intracellular markers of interest in the myeloid compartment. Antibodies were purchased pre-conjugated or conjugated in house based on manufacturer guidelines (Standard BioTools). Samples were barcoded with palladium isotopes prior to staining to reduce the impact of batch effect, and were then analyzed on a first generation CyTOF system (Standard BioTools). Following background attenuation and singlet gating in FlowJo (BD), the CATALYST R package enabled deconvolution and visualization of single cell data, including dimensional reduction techniques (tSNE) and clustering of all cell populations. An iterative approach was taken to determine a stable configuration of clusters across sample populations to protect from over-fragmentation of data, further informed by K-means clustering using the elbow method. Heatmaps were then generated based on median scaled expression of markers contained in each cluster.
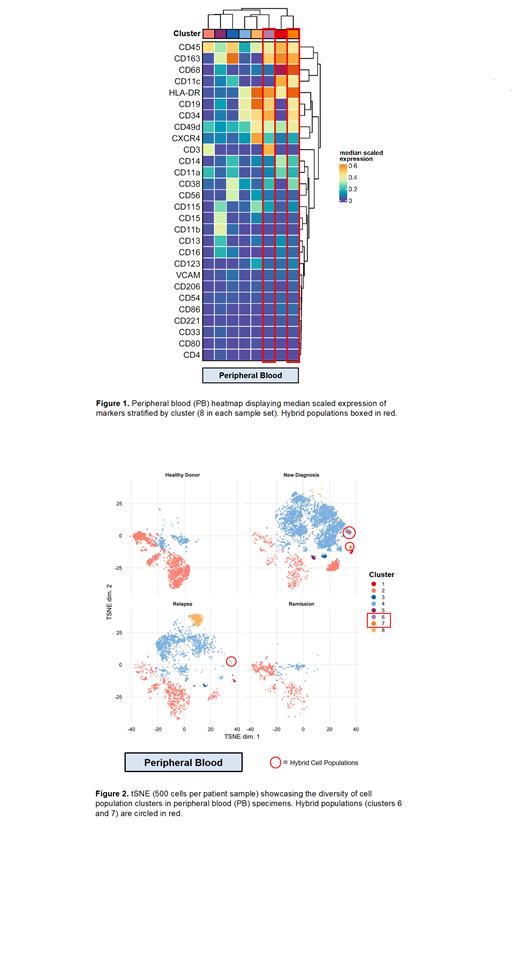
Results: The CyTOF panel identified several different myeloid cell populations in newly diagnosed, relapsed, and remission samples in both PB and BM compared to HDs. Most interestingly, quantifiable levels of a cluster of cells co-expressing CD19, CD3 and CD163 as well as a second cluster of cells expressing CD19 and CD163 were detected in newly diagnosed and relapsed patients, but not in HDs. A single patient with the largest proportion of these cell populations in their PB at the time of relapse presented with an aggressive leukemia variant and relapsed multiple times prior to succumbing to illness. On investigation of BM aspirates, the triple positive hybrid population was found in a similar distribution amongst the newly diagnosed and relapsed patients, as well as affecting one patient who achieved remission. This patient also had a highly aggressive leukemia that was refractory to chemotherapy and immunotherapy, and only achieved this remission following CAR-T cell therapy. She subsequently underwent stem cell transplant and is in remission.
Conclusions: Our findings add to the literature highlighting the utility of high parameter single cell capture techniques in detecting rare cell populations in the leukemic TME. Further, we identified two unique hybrid cell populations that are reliably detected across different disease stages in patients with B-ALL. Importantly, these populations persisted following stringent gating protocols to rule out doublets and debris. Although our sample size was too small to demonstrate a prognostic impact of these hybrid cell populations in patients with B-ALL, the identification of these populations alone is a significant finding. Further investigation and patient sample accrual are warranted to better understand the role of these cell types in tumor biology, which may pave the way towards new prognostic models that can leverage well-established immune monitoring techniques.
Disclosures
Lind:Kronos Bio: Research Funding. Leonard:Pfizer: Consultancy; Adaptive Biotechnologies: Consultancy, Membership on an entity's Board of Directors or advisory committees, Other: Travel, accommodations, expenses; Takeda: Consultancy; Kite/Gilead: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal